The morning sessions (starting at 8:30 am) will be devoted to lectures, and the afternoon sessions to practicals.
On Monday afternoon, each participant will provide a brief overview of their research topic and interests.
Program overview
- Sunday 17th: Arrival
- Monday 18th: Deep learning for scientific applications, Julien Mairal (Inria, France)
- Tuesday 19th: AlphaFold: A Case Study in Deep Learning for Protein Structure Prediction, Paulyna Magaña and Maxim Tsenkov (EMBL-EBI, UK)
- Wednesday 20th: Design of functional biomolecules using deep generative models, Alisa Khramushin and Ilia Igashov (Correia lab, EPFL, Switzerland)
- Thursday 21st: Artificial Intelligence deciphers the code of life written in proteins, Burkhard Rost and Tobias Senoner (TUM, Germany)
- Friday 22nd: Deep Generative Models for Sampling at Equilibrium, Marylou Gabrié (CMAP, École Polytechnique, France)
Monday 18th: Deep learning for scientific applications
Lecturer: Julien Mairal (Inria Grenoble, France)
Contents:
- Brief introduction to common deep learning models
- Beyond black boxes with physics-informed machine learning
- Self-supervised learning in computer vision and ``foundation'' models
- Current challenges about self-supervised learning for molecular representation
Link to the slides
Tuesday 19th: AlphaFold: A Case Study in Deep Learning for Protein Structure Prediction
Lecturer: Paulyna Magaña and Maxim Tsenkov (EMBL-EBI, UK)
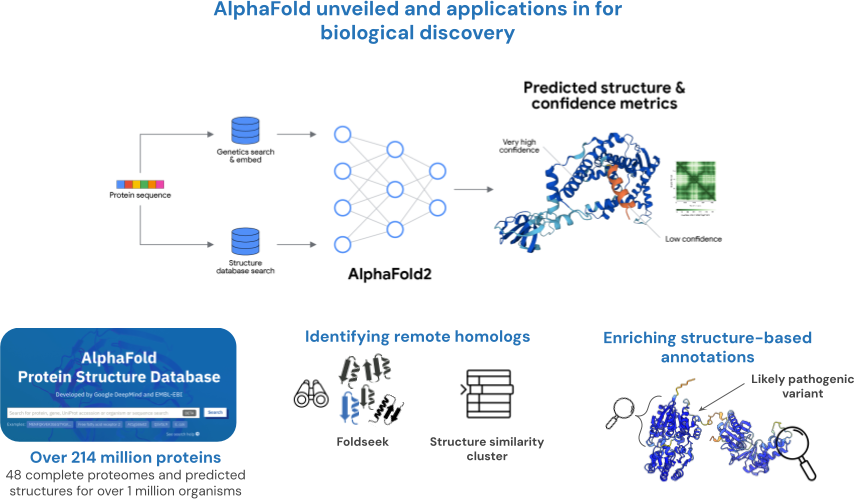
Lecture contents:
- AlphaFold: A Deep Learning "Solution".
- Impact of AlphaFold on biological research.
- Limitations and future directions of AlphaFold.
- Enriching predicted structures from AlphaFold.
Practical session:
- Sampling protein structure predictions.
- Identifying remote homologous.
- Evaluating and enriching predicted structures from AlphaFold DB.
Wednesday 20th: Design of functional biomolecules using deep generative models
Lecturers: Alisa Khramushin and Ilia Igashov (Correia lab, EPFL, Switzerland)
(image taken from Khakzad, H., Igashov, I., Schneuing, A., Goverde, C., Bronstein, M. and Correia, B., 2023. A new age in protein design empowered by deep learning. Cell Systems, 14(11), pp.925-939)
Lecture contents:
- Protein design: existing algorithms, challenges and current developments
- The theory of deep generative models for generation of new biomolecules: score-based models and flow-matching
- Generative models in protein design and drug discovery
Practical session:
- RoseTTAFold diffusion for protein binder design
- Sequence design with ProteinMPNN
- Analysis of the results using AF
- Structure-based drug generation with DiffSBDD
Thursday 21st: Artificial Intelligence deciphers the code of life written in proteins
Lecturers: Burkhard Rost and Tobias Senoner (TUM, Germany)
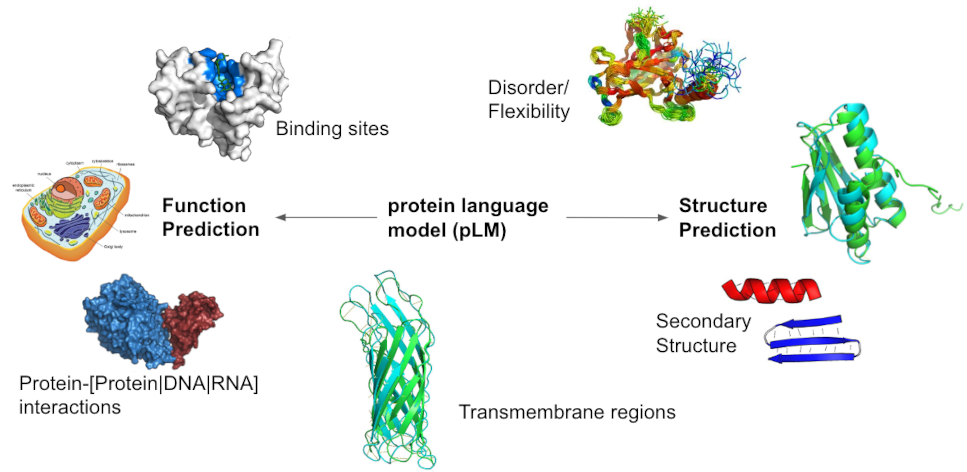
Contents:
- Historical reliance on expert features in AI for protein prediction
- Transition to embeddings from large language models in AI
- Role and pitfalls of protein embeddings in protein prediction
- Methods for exploring and visualizing protein embeddings
- Current limitations and future strategies for protein embeddings
Friday 22nd: Deep Generative Models for Sampling at Equilibrium
Lecturer: Marylou Gabrié (CMAP, École Polytechnique, France)
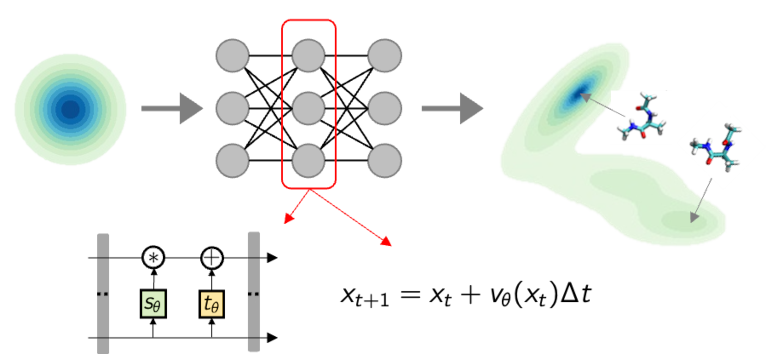
Contents:
- Transport based generative models: discrete and continuous designs including geometric equivariances
- Training and sampling pipelines with generative models
- Revisiting enhanced samplers based on collective variables with normalising flows






